Process#
The Process class is used to define dynamic changes of model parameters or flow sheet connections.
To instantiate a Process, a FlowSheet needs to be passed as argument, as well as a string to name that process.
For this guide, a Batch Elution Flow Sheet FlowSheet is used.
For more information on the FlowSheet, refer to Flow Sheet.
from CADETProcess.processModel import Process
process = Process(flow_sheet, 'batch_elution')
After instantiation, it is important to also set the overall duration of the process.
Since CADET-Process is also designed for cyclic processes (see Cyclic Stationarity), the corresponding attribute is called cycle_time.
process.cycle_time = 10 # s
The add_event() method requires the following arguments:
name: Name of the event.parameter_path: Path of the parameter that is changed in dot notation. E.g. to specify theflow_rateparameter ofInletunit namedeluent, of theFlowSheetwould read asflow_sheet.eluent.flow_rate.state: Value of the attribute that is changed at Event execution.time: Time at which the event is executed in \(s\).
For example, to add an event called inject_on which should modify flow_rate of unit feed to value 1 at time 0, and an event called inject_off which sets the flow rate back to 0 at \(t = 1~s\), specify the following:
process.add_event('inject_on', 'flow_sheet.feed.flow_rate', 1, 0)
process.add_event('inject_off', 'flow_sheet.feed.flow_rate', 0, 1)
If the event shall only modify a single entry of an array, an additional indices argument can be added.
For example, the following will only modify the concentration of the first component.
process.add_event('conc_high', 'flow_sheet.feed.c', 1, 0, indices=0)
process.add_event('conc_low', 'flow_sheet.feed.c', 0, 1, indices=0)
All events can are stored in the events attribute.
To visualize the trajectory of the parameter state over the entire cycle, the Process provides a plot_events() method.
_ = process.plot_events()
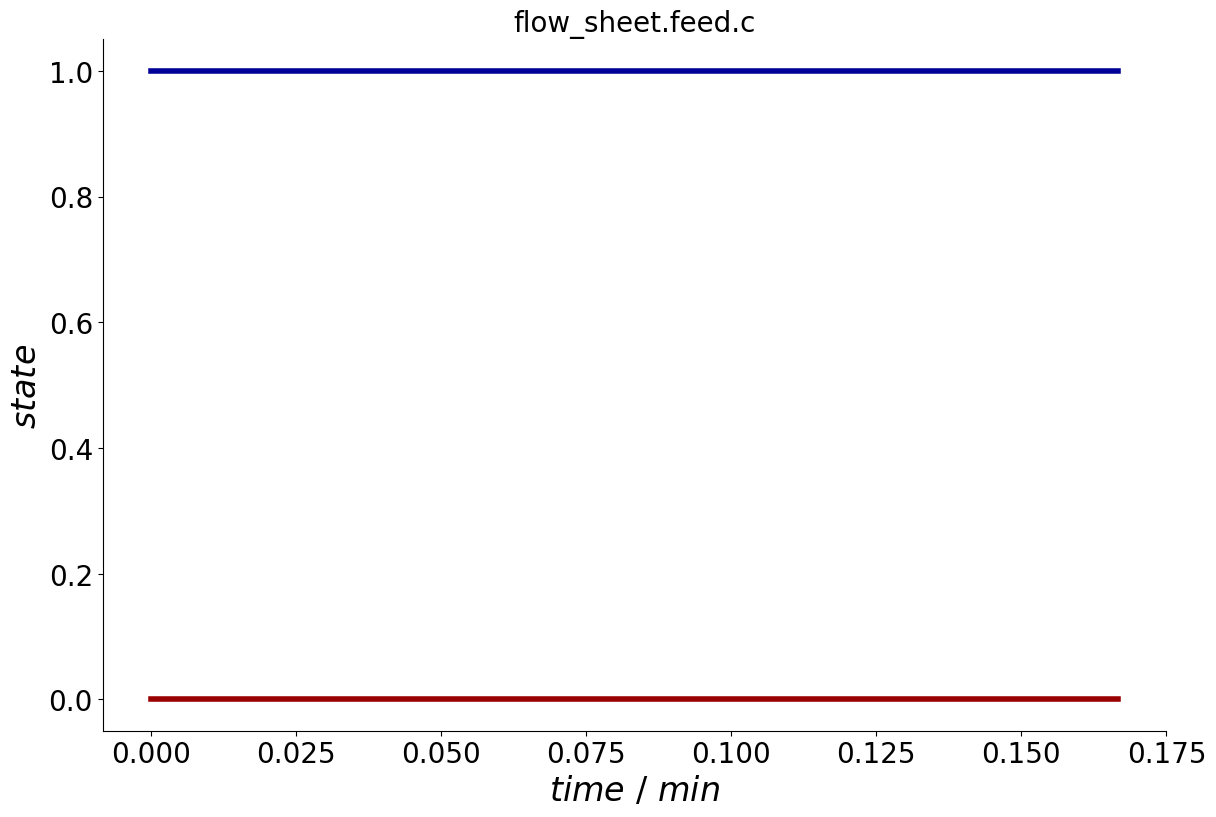
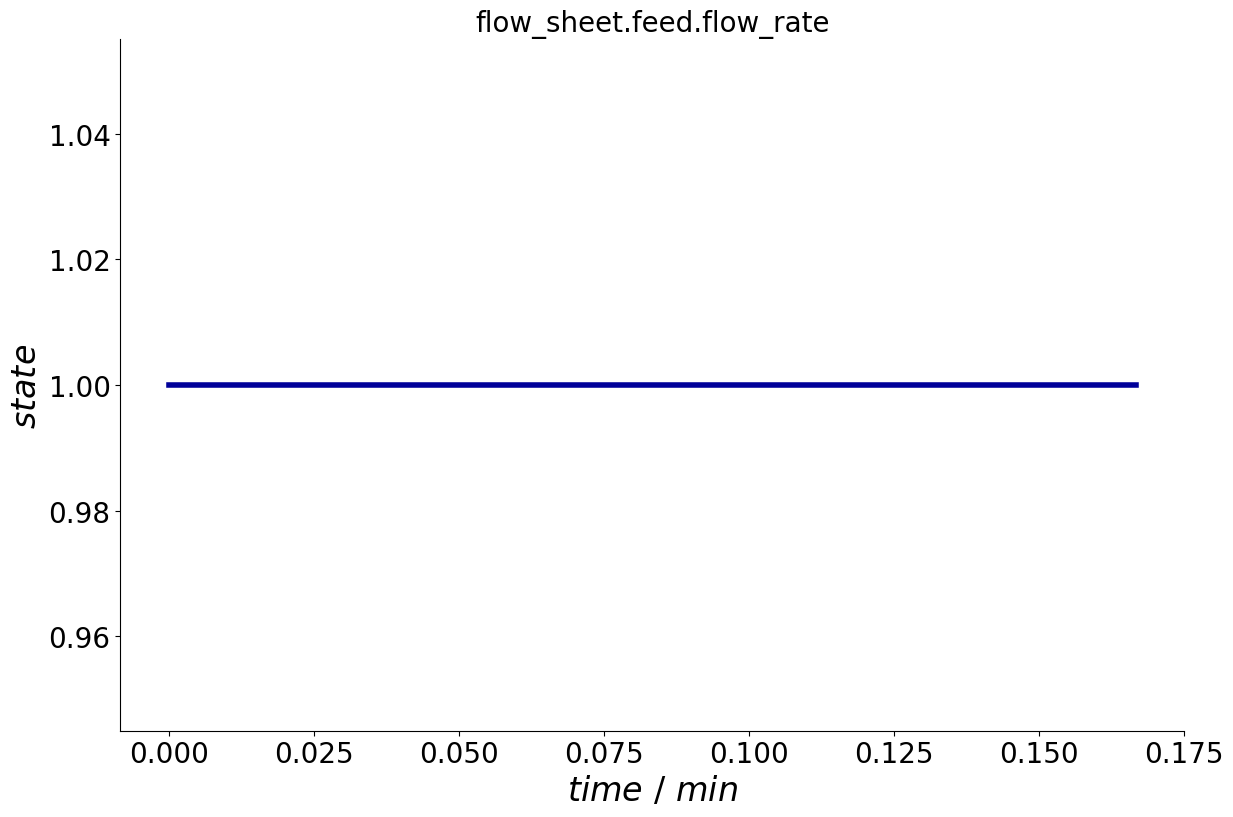
Event Dependencies#
In order to reduce the complexity of process configurations, Event dependencies can be specified that define the time at which an Event occurs as a function of other Event times.
Especially for more advanced processes, this reduces the degrees of freedom and facilitates the overall handiness.
For this purpose, Durations can also be defined to describe the time between the execution of two Events.
The time of a dependent event is calculated using the following equation
where \(n_{dep}\) denotes the number of dependencies of a given Event, \(\lambda_i\) denotes a linear factor, and \(f_i\) is some transform function.
To add a dependency in CADET-Process, use the add_event_dependency() method of the Process class which requires the following arguments:
dependent_event: Name of the event whose value will depend on other events.independent_events: List of other events on which event dependsfactors: List with factors for linear combination of dependencies.
Consider the batch elution process (see here).
Events of batch elution process.#
Here, every time the feed is switched on, the elution buffer should be switched off and vice versa.
process.add_event('feed_on', 'flow_sheet.feed.flow_rate', Q)
process.add_event('feed_off', 'flow_sheet.feed.flow_rate', 0.0)
process.add_event('eluent_off', 'flow_sheet.eluent.flow_rate', 0.0)
process.add_event_dependency('eluent_off', ['feed_on'])
Alternatively, the dependencies can also already be added in the add_event() method when creating the Event in the first place.
process.add_event('eluent_on', 'flow_sheet.eluent.flow_rate', Q, dependencies=['feed_off'])
Now, only one of the event times needs to be adjusted, e.g. in a process optimization setting.
process.feed_off.time = 10
print(f'feed_off: {process.feed_off.time}')
print(f'eluent_on: {process.eluent_on.time}')
feed_off: 10.0
eluent_on: 10.0
For a more complex scenario refer to SSR process.
Specifying Indices of Multidimensional Parameters for Events#
Many model parameters in CADET-Process are not scalar and their size may depend on the number of components, binding sites, reactions, etc. CADET-Process provides some functionality to manage these sized parameters when adding events.
For this tutorial, consider the following process model:
Show code cell content
import copy
import numpy as np
from CADETProcess.processModel import ComponentSystem
component_system = ComponentSystem(2)
from CADETProcess.processModel import Inlet
inlet = Inlet(component_system, 'inlet')
from CADETProcess.processModel import Inlet, LumpedRateModelWithPores, Outlet
inlet = Inlet(component_system, name='inlet')
column = LumpedRateModelWithPores(component_system, 'column')
outlet = Outlet(component_system, 'outlet')
from CADETProcess.processModel import FlowSheet, Process
flow_sheet = FlowSheet(component_system)
flow_sheet.add_unit(inlet)
flow_sheet.add_unit(column)
flow_sheet.add_unit(outlet)
flow_sheet.add_connection(inlet, column)
flow_sheet.add_connection(column, outlet)
def setup_process():
process = Process(flow_sheet, 'Demo Indices')
process.cycle_time = 10
return process
Specifying Indices for 1D Arrays#
Consider a 1D parameter, such as the film_diffusion coefficient of the LumpedRateModelWithPores.
If no indices are provided, all entries of the parameter are set to the same value,
E.g., to change the value of all film diffusion coefficients to the value 1e-6 at time=0, add the following:
process = setup_process()
process.add_event(
'film_diffusion_all', 'flow_sheet.column.film_diffusion', [1e-6, 1e-6], time=0
)
print(column.film_diffusion)
[1e-06, 1e-06]
To add an event for a single entry in a 1D list / array, add an indices argument that specifies the index of the entry to be modified.
E.g., to only change the value of the first component’s film diffusion coefficient to 1e-7 at time=1, add indices=0.
process.add_event(
'film_diffusion_index_0', 'flow_sheet.column.film_diffusion', 1e-7, indices=0, time=1
)
print(column.film_diffusion)
[1e-07, 1e-06]
It is also possible to specify multiple indices at once.
The length of indices must match the length of the provided states.
Also, the order of the indices is mapped to the order of the state entries.
process.add_event(
'film_diffusion_index_10', 'flow_sheet.column.film_diffusion', [2e-7, 3e-7], indices=[1, 0], time=2
)
print(column.film_diffusion)
[3e-07, 2e-07]
Specifying Indices for 1D Polynomial Parameters#
As mentioned here, polynomial parameters are a bit special with regard to their setter methods.
As with regular 1D parameters, it is possible to specify all state values at once.
To demonstrate this, consider the flow_rate attribute of the inlet unit operation.
process = setup_process()
process.add_event(
'flow_rate_all', 'flow_sheet.inlet.flow_rate', [0, 1, 2, 3], time=0
)
print(inlet.flow_rate)
[0. 1. 2. 3.]
Also events that only modify a single entry are equivalent:
process.add_event(
'flow_rate_index_0', 'flow_sheet.inlet.flow_rate', 1, indices=0, time=1
)
print(inlet.flow_rate)
[1. 1. 2. 3.]
However, if no indices are passed, the event acts as if the state is set directly to the parameter.
Consequently, setting the state to 1 would automatically fill missing polynomial coefficients and set them all to 0.
process.add_event(
'flow_rate_fill_all', 'flow_sheet.inlet.flow_rate', 1, time=2
)
print(inlet.flow_rate)
[1. 0. 0. 0.]
Adding a subset of the coefficients as list also works as expected:
process.add_event(
'flow_rate_fill_subset', 'flow_sheet.inlet.flow_rate', [2, 1], time=3
)
print(inlet.flow_rate)
[2. 1. 0. 0.]
Specifying Indices for Multidimensional (Polynomial) Parameters#
The process for adding events for multidimensional parameters is similar.
For this purpose, consider the concentration c of the inlet unit operation (which also happens to be polynomial).
Here, the parameter is fully specified in the event:
process = setup_process()
evt = process.add_event(
'c_all', 'flow_sheet.inlet.c',
[[0, 1, 2, 3], [4, 5, 6, 7]],
time=0
)
print(inlet.c)
[[0. 1. 2. 3.]
[4. 5. 6. 7.]]
To add an event that only modify a single entry, note that indices must now be a tuple containing the index of every dimension of the array.
evt = process.add_event(
'c_single', 'flow_sheet.inlet.c', 1, indices=(0, 0), time=1
)
print(inlet.c)
[[1. 1. 2. 3.]
[4. 5. 6. 7.]]
To add multiple indices, make sure to pass a list of tuples.
evt = process.add_event(
'c_multi', 'flow_sheet.inlet.c', [2, 3], indices=[(0, 0), (1, 1)], time=2
)
print(inlet.c)
[[2. 1. 2. 3.]
[4. 3. 6. 7.]]
Slicing is also possible using np.s_.
For more information, refer to this post.
E.g. to add an event that modifies all entries of the first row:
evt = process.add_event(
'c_row', 'flow_sheet.inlet.c', 1, indices=np.s_[0, :], time=3
)
print(inlet.c)
[[1. 1. 1. 1.]
[4. 3. 6. 7.]]
As with 1D polynomials, leveraging the setter convenience function is also possible for polynomial parameters.
E.g. to set the first entry to a constant 2 and the second to value where the constant coefficient is 1 and the linear coefficient is 2 specify the following as state: [2, [1, 2]].
Note, this only works when no indices are provided.
evt = process.add_event(
'c_poly', 'flow_sheet.inlet.c', [2, [1, 2]], time=4
)
print(inlet.c)
[[2. 0. 0. 0.]
[1. 2. 0. 0.]]
Generating Inlet Profiles from Existing Data#
In some cases, it is desirable to use an existing concentration profile as inlet profile (e.g. the output of some upstream process). CADET-Process provides a method to automatically fit a piecewise cubic polynomial to a given profile and add the corresponding events to a specific inlet.
E.g. consider this (arbitrary) sinusoidal profile:
Text(0.5, 0, 'time / min')
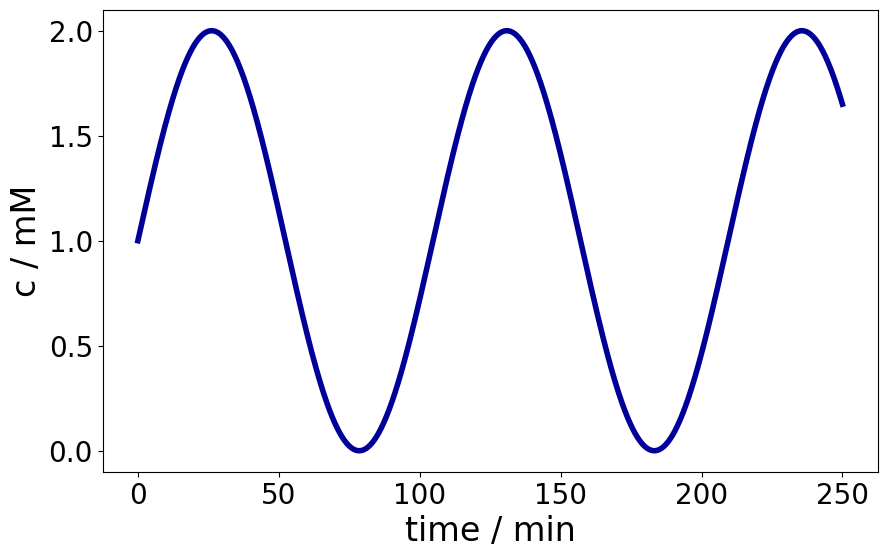
Consider that the time points are stored in a variable t, and the concentration in the variable c.
To add the profile to the unit called inlet, use the add_concentration_profile() method of the Process with the following arguments:
unit_operation: Inlet unit operationtime: Time pointsc: Concentration valuescomponents: List of component species to which the concentration profile shall be added. If-1, the same profile is set for all components.
process.add_concentration_profile('inlet', time, c, components=-1)
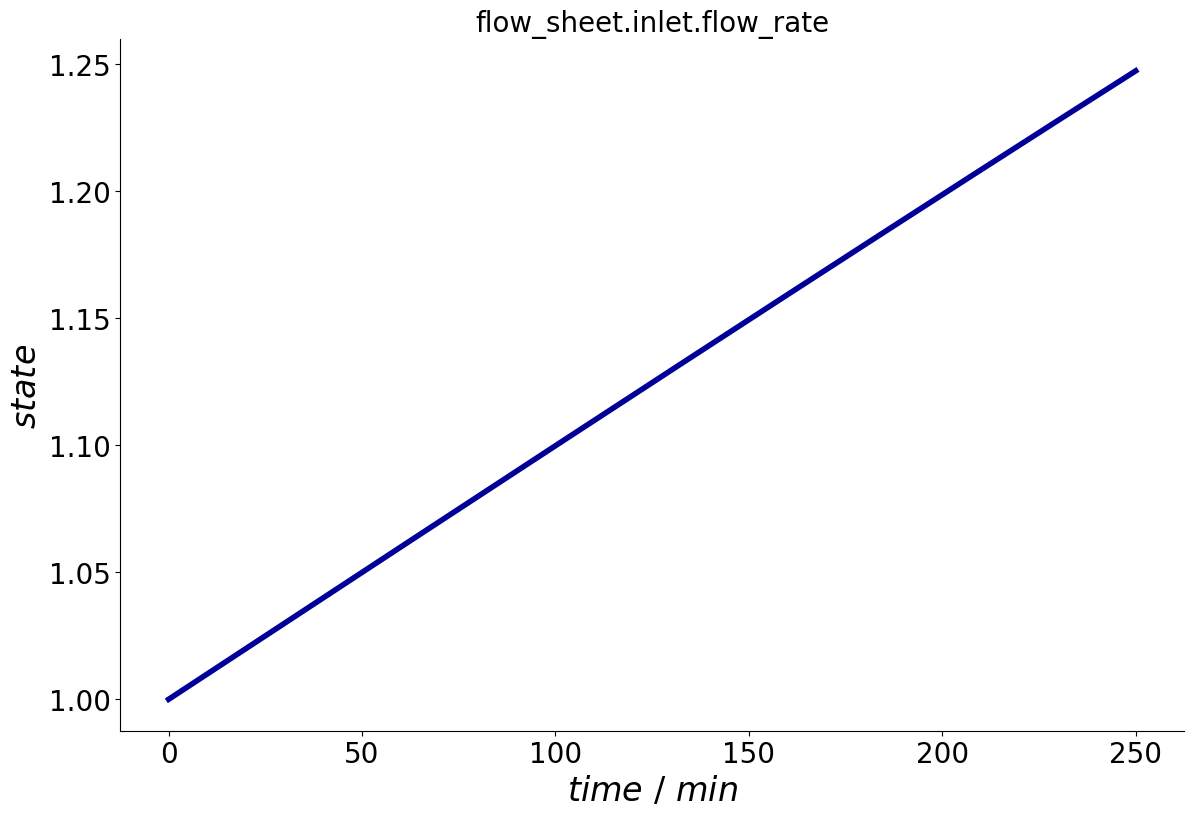
When calling the plot_events() method, the concentration profile can now be seen.
_ = process.plot_events()
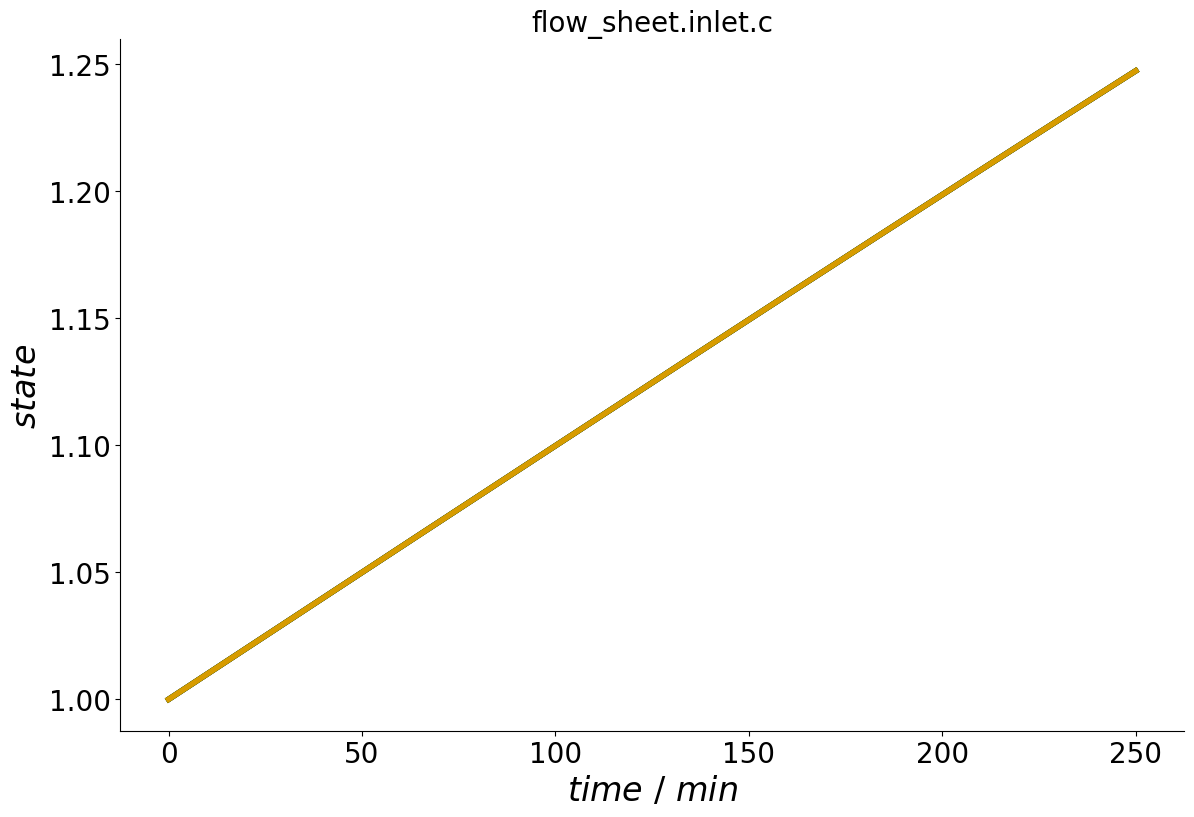
Similarly, flow rate profiles can be added to Inlets and Cstrs.
Note that for obvious reasons, the component index is omitted.
process.add_flow_rate_profile('inlet', time, c)
_ = process.plot_events()
Parameter Sensitivities#
Warning
This functionality is still work in progress. Changes in the interface are to be expected.
Parameter sensitivities \(s = \partial y / \partial p\) of a solution \(y\) with respect to some parameter \(p\) are required for various tasks, for example, parameter estimation, process design, and process analysis.
The CADET simulator implements the forward sensitivity approach which creates a linear companion DAE for each sensitive parameter.
To add a parameter sensitivity to the process, use the add_parameter_sensitivity() method.
The first argument is the parameter path in dot notation.
Since currently parameter sensitivities cannot be computed for output_states or event times, it is not required to specify flow_sheet in the path.
Optionally, a name can be provided.
If none is provided, the parameter path is used.
process.add_parameter_sensitivity('column.total_porosity')
For parameters that are specific to a component, reaction, bound index, etc., this must also be specified in the method.
process.add_parameter_sensitivity(
'feed.c',
components=['A'],
polynomial_coefficients=0,
section_indices=0,
)
Multiple parameters can also be linearly combined.
For each of the arguments, a list of parameter needs to be passed.
In this case, a name also must be provided.
For more information on the computed sensitivities, refer to Simulation Results.
