Product Fractionation#
Key information for evaluating the separation performance of a chromatographic process are the amounts of the target components in the collected product fractions. To define corresponding fractionation intervals, the chromatograms, i.e., the concentration profiles \(c_{i,k}\left(t\right)\) at the outlet(s) of the process must be evaluated. In a strict sense, a chromatogram is only given at the outlet of a single column. Note that here this term is used more generally for the concentration profiles at the outlets of a flow sheet, which only accounts for material leaving the process. The times for the start, \(t_{start, j}\), and the end, \(t_{end, j}\), of a product fraction \(j\) have to be chosen such that constraints on product purity are met. It is important to note, that in advanced chromatographic process configurations, outlet chromatograms can be much more complex than the example shown below and that multiple sections of the chromatogram may represent suitable fractions \(j\) for collecting one target component \(i\). Moreover, flow sheets can have multiple outlets \(k\) that have to be fractionated simultaneously. Also, the volumetric flow rate \(Q_k\) at the outlets may depend on time and needs to be considered in the integral. These aspects are considered by defining the total product amount of a component \(i\) as
where \(n_{frac, k}^{i}\) is the number of fractions considered for component \(i\) in chromatogram \(k\), and \(n_{chrom}\) is the number of chromatograms that is evaluated.
Further performance criteria typically used for evaluation and optimization of chromatographic performance are the specific productivity, \(PR_i\), the recovery yield, \(Y_i\), and the specific solvent consumption, \(EC_i\), which all depend on the product amounts:
with \(V_{solid}\) being the volume of stationary phase, \(V_{solvent}\) that of the solvent introduced during a cycle with duration \(\Delta t_{cycle}\), and \(m_{feed}\) the injected amount of mixture to be separated. Multiple Inlets can be considered for the amounts of consumed feed and solvent,
For the cumulative product purities \(PU_i\) holds
where \(n_{comp}\) is the number of mixture components and \(m_{l}^{i}\) is the mass of component \(l\) in target fraction \(i\).
In CADET-Process, the fractionation module provides methods to calculate these performance indicators.
Fractionator#
The Fractionator allows slicing the solution and pool fractions for the individual components.
It enables evaluating multiple chromatograms at once and multiple fractions per component per chromatogram.
The most basic strategy is to manually set all fractionation times manually. To demonstrate the strategy, consider a simplebatch-elution example.
To enable the calculation of the process parameters, it is necessary to specify which of the inlets should be considered for the feed and eluent consumption. Moreover, the outlet(s) which are used for evaluation need to be defined.
flow_sheet.add_feed_inlet('feed')
flow_sheet.add_eluent_inlet('eluent')
flow_sheet.add_product_outlet('outlet')
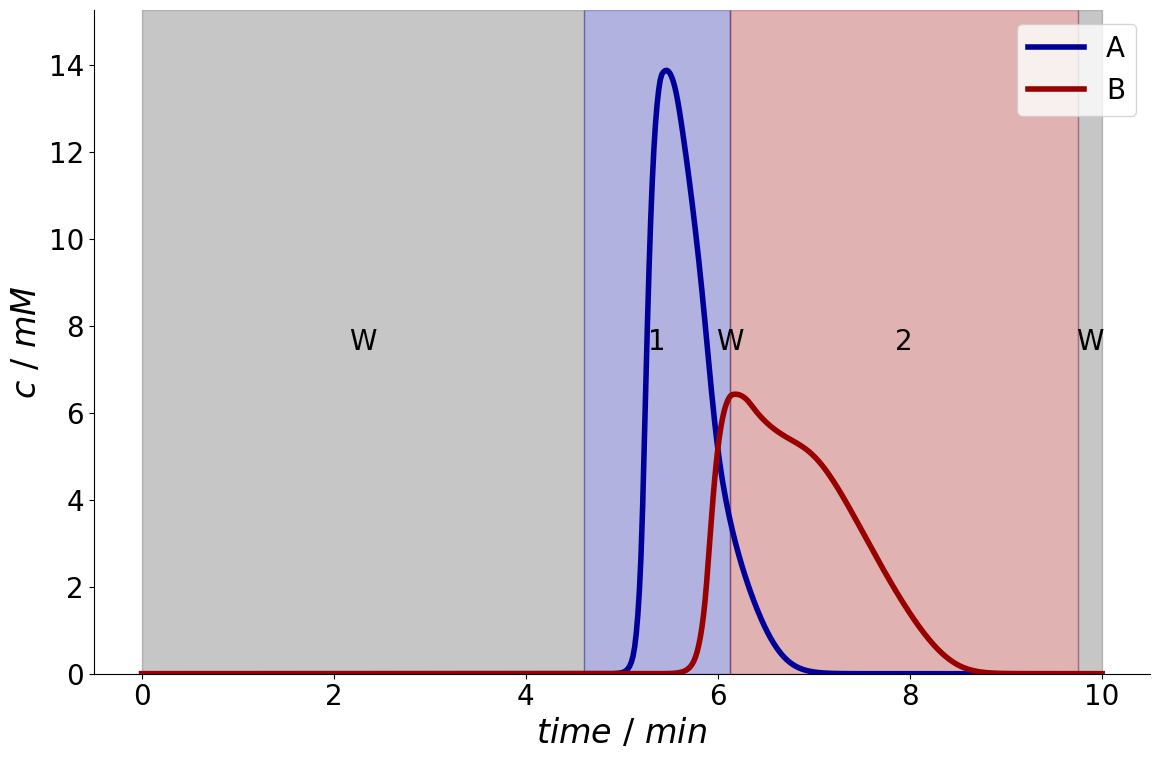
For reference, this is the chromatogram at the outlet that needs to be fractionated:
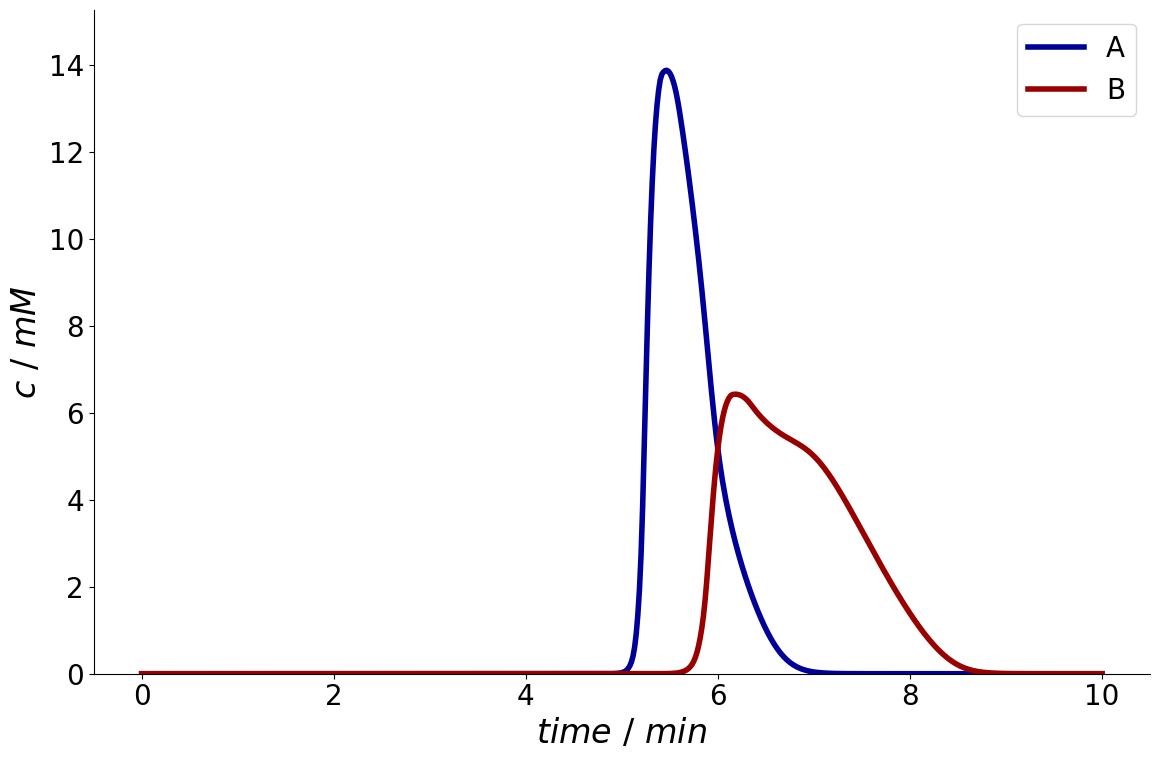
Concentration profile at column outlet.#
After import, the Fractionator is instantiated with the simulation results.
from CADETProcess.fractionation import Fractionator
fractionator = Fractionator(simulation_results)
To add a fractionation event, the following arguments need to be provided:
event_name: Name of the event.target: Pool to which fraction is added.-1indicates waste.time: Time of the eventchromatogram: Name of the chromatogram. Optional if only one outlet is set asproduct_outlet.
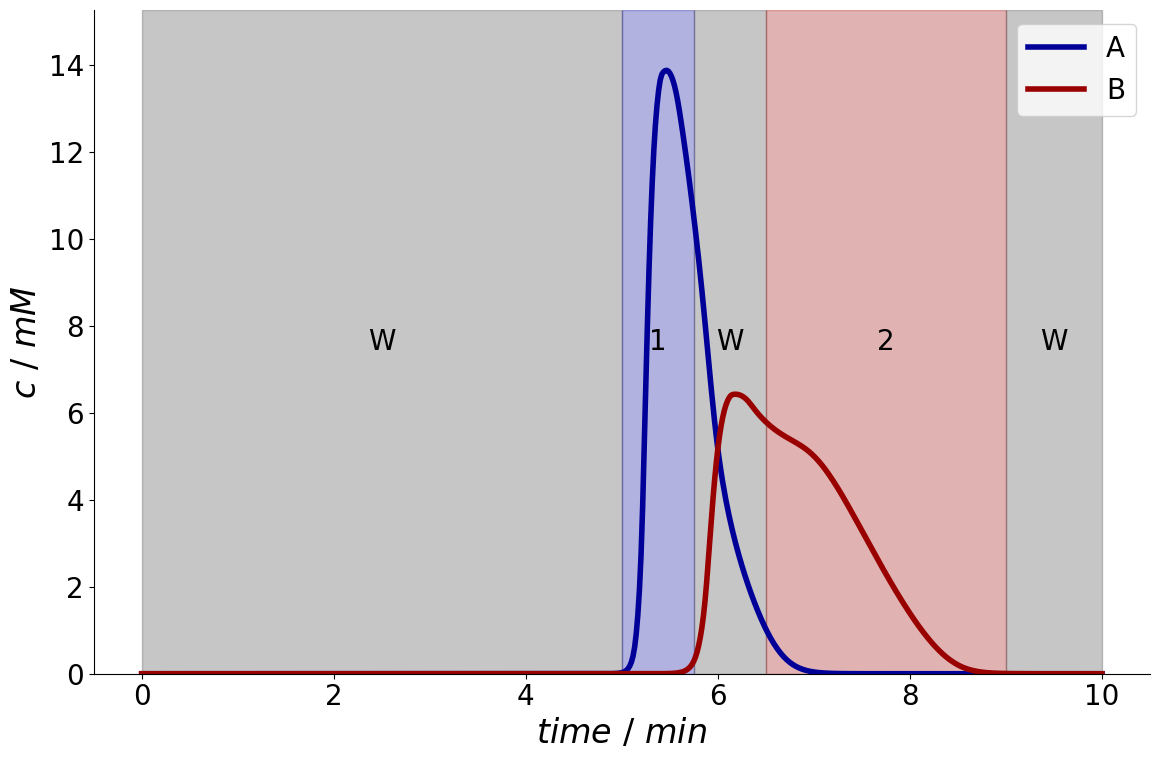
Here, component \(A\) seems to have sufficient purity between \(5 \colon 00~min\) and \(5 \colon 45~min\) and component \(B\) between \(6 \colon 30~min\) and \(9 \colon 00~min\).
fractionator.add_fractionation_event('start_A', 0, 5*60, 'outlet')
fractionator.add_fractionation_event('end_A', -1, 5.75*60)
fractionator.add_fractionation_event('start_B', 1, 6.5*60)
fractionator.add_fractionation_event('end_B', -1, 9*60)
The Performance object of the Fractionator contains the parameters:
print(fractionator.performance)
Performance(mass=array([0.00039127, 0.00038088]), concentration=array([8.69481795, 2.53920449]), purity=array([0.99721983, 0.97224987]), recovery=array([0.65211135, 0.63480112]), productivity=array([0.00800824, 0.00779566]), eluent_consumption=array([0.72456816, 0.70533458]) mass_balance_difference=array([-1.46459000e-10, 9.08286074e-10]))
With these fractionation times, the both component fractions reach a purity of \(99.7~\%\), and \(97.2~\%\) respectively. The recovery yields are \(65.2~\%\) and \(63.4~\%\).
The chromatogram can be plotted with the fraction times overlaid:
_ = fractionator.plot_fraction_signal()
Optimization of Fractionation Times#
The fractionation module also provides a method to set up an OptimizationProblem which automatically determines optimal cut times.
For every component, different purity requirements can be specified, and any function may be applied as objective.
For the objective and constraint functions, fractions are pooled from all Outlets of the FlowSheet (see equations (1) and (7)) that have been marked as product_outlet.
For more information about configuring the FlowSheet, refer to Flow Sheet.
As initial values for the optimization, areas of the chromatogram with sufficient local purity are identified, i.e., intervals where \(PU_i(t)=c_i(t)/\sum_j c_j(t)\geq PU_{min,i}\) [2].
These initial intervals are then expanded by the optimizer towards regions of lower purity while meeting the cumulative purity constraints.
In the current implementation, COBYLA [3] of the SciPy [4] library is used as optimizer.
Yet, any other solver or heuristic algorithm may be used.
from CADETProcess.fractionation import FractionationOptimizer
fractionation_optimizer = FractionationOptimizer()
By default, the mass of the components is maximized under purity constraints. However, other objective functions can be used.
To automatically optimize the fractionation times, pass the simulation results to the optimize_fractionation() method.
Depending on the separation problem at hand, different purity requirements can be specified.
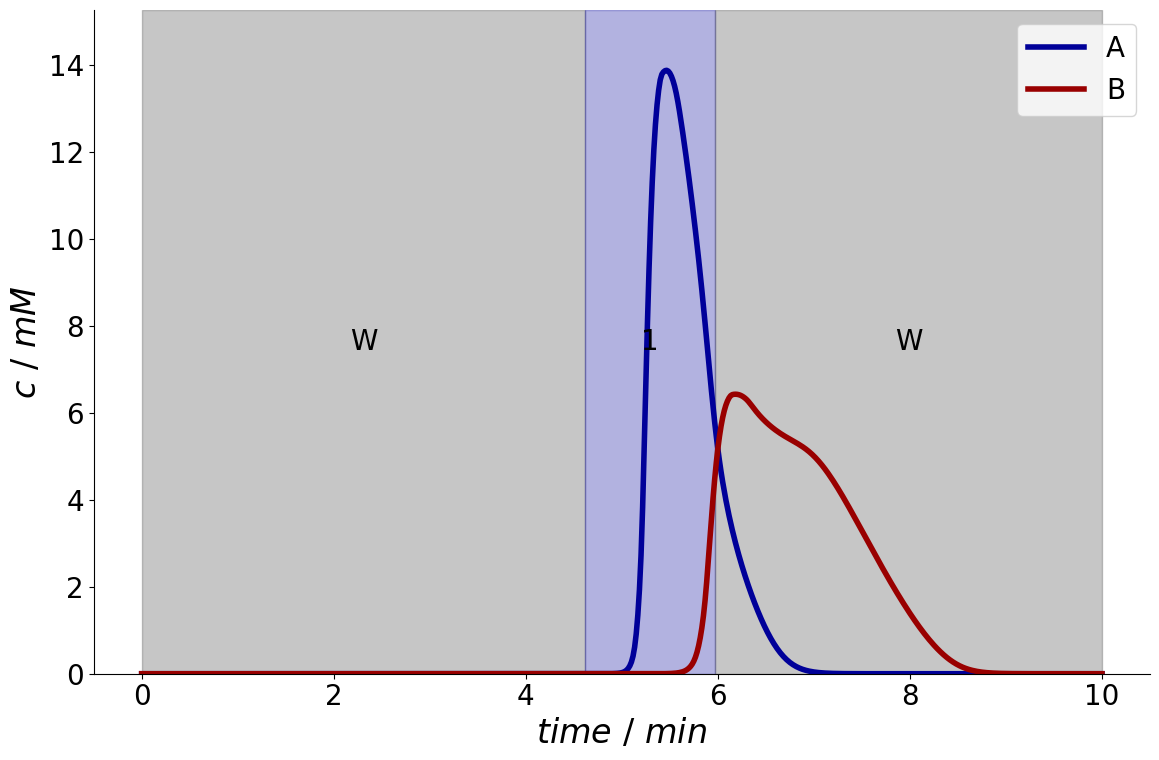
For example, here only the first component is relevant, and requires a purity \(\ge 95~\%\):
fractionator = fractionation_optimizer.optimize_fractionation(simulation_results, purity_required=[0.95, 0])
The results are stored in a Performance object.
print(fractionator.performance)
Performance(mass=array([0.00049949, 0. ]), concentration=array([6.16946937, 0. ]), purity=array([0.94986252, 0. ]), recovery=array([0.83247844, 0. ]), productivity=array([0.01022323, 0. ]), eluent_consumption=array([0.92497605, 0. ]) mass_balance_difference=array([-9.72000556e-11, 1.10215308e-09]))
The chromatogram can also be plotted with the fraction times overlaid:
_ = fractionator.plot_fraction_signal()
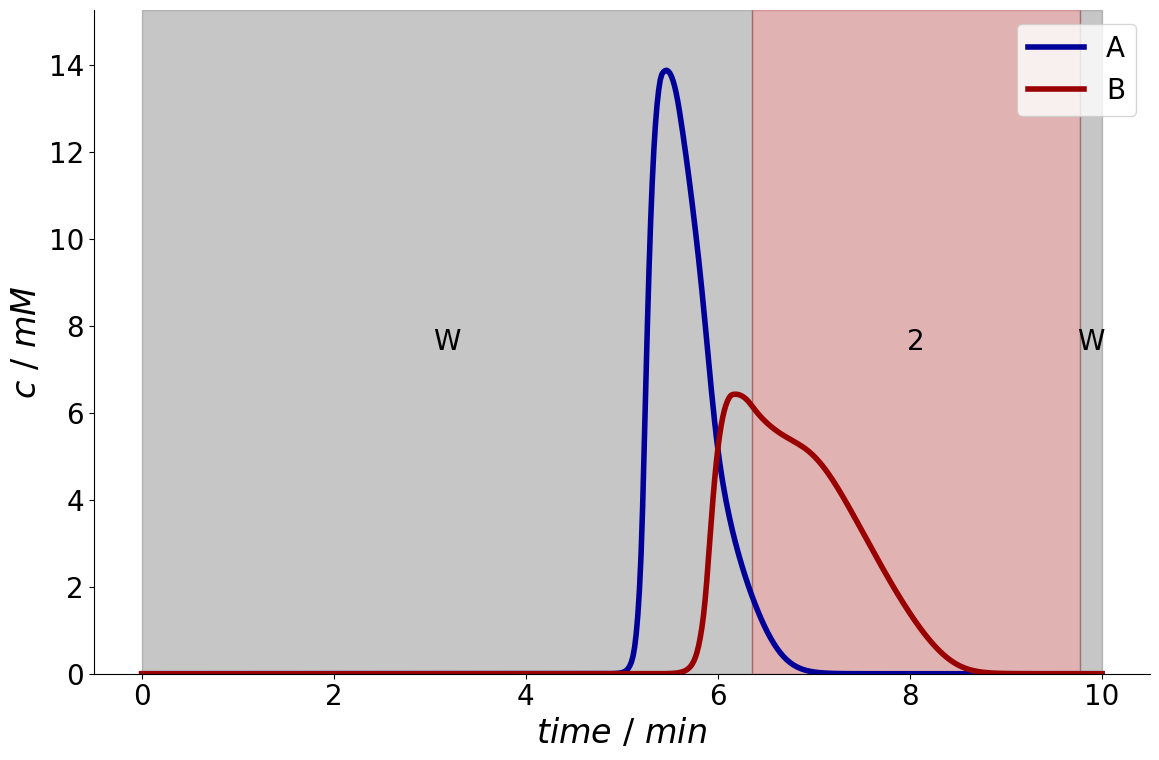
For comparison, this is the results if only the second component is relevant:
fractionator = fractionation_optimizer.optimize_fractionation(simulation_results, purity_required=[0, 0.95])
print(fractionator.performance)
_ = fractionator.plot_fraction_signal()
Performance(mass=array([0. , 0.00043243]), concentration=array([0. , 2.11333831]), purity=array([0. , 0.94977569]), recovery=array([0. , 0.72071895]), productivity=array([0. , 0.00885077]), eluent_consumption=array([0. , 0.80079883]) mass_balance_difference=array([1.90933729e-10, 9.54821213e-11]))
But of course, also both components can be valuable. Here, the required purity is also reduced to demonstrate that overlapping fractions are automatically avoided by internally introducing linear constraints.
fractionator = fractionation_optimizer.optimize_fractionation(simulation_results, purity_required=[0.8, 0.8])
print(fractionator.performance)
_ = fractionator.plot_fraction_signal()
Performance(mass=array([0.00054079, 0.00052124]), concentration=array([5.96025791, 2.39228225]), purity=array([0.87288289, 0.8979969 ]), recovery=array([0.90132006, 0.86874101]), productivity=array([0.01106864, 0.01066855]), eluent_consumption=array([1.00146673, 0.96526779]) mass_balance_difference=array([-3.87966905e-12, -3.58058325e-10]))
To set an alternative objective, a function needs to be passed that takes a Performance as an input.
In this example, not only the total mass is considered important but also the concentration of the fraction.
As previously mentioned, COBYLA only handles single objectives.
Hence, a RankedPerformance is used which transforms the Performance object by adding a weight \(w_i\) to each component.
It is important to remember that by default, objectives are minimized.
To register a function that is to be maximized, add the minimize=False flag.
Also, the number of objectives the function returns needs to be specified.
from CADETProcess.performance import RankedPerformance
ranking = [1, 1]
def alternative_objective(performance):
performance = RankedPerformance(performance, ranking)
return performance.mass * performance.concentration
fractionator = fractionation_optimizer.optimize_fractionation(
simulation_results, purity_required=[0.95, 0.95],
obj_fun=alternative_objective,
n_objectives=1,
minimize=False,
)
print(fractionator.performance)
_ = fractionator.plot_fraction_signal()
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[15], line 7
4 performance = RankedPerformance(performance, ranking)
5 return performance.mass * performance.concentration
----> 7 fractionator = fractionation_optimizer.optimize_fractionation(
8 simulation_results, purity_required=[0.95, 0.95],
9 obj_fun=alternative_objective,
10 n_objectives=1,
11 minimize=False,
12 )
14 print(fractionator.performance)
15 _ = fractionator.plot_fraction_signal()
File ~/checkouts/readthedocs.org/user_builds/cadet-process/conda/latest/lib/python3.11/site-packages/CADETProcess/fractionation/fractionationOptimizer.py:368, in FractionationOptimizer.optimize_fractionation(self, simulation_results, purity_required, components, use_total_concentration_components, ranking, obj_fun, n_objectives, bad_metrics, minimize, allow_empty_fractions, ignore_failed, return_optimization_results, save_results)
358 simulation_results.process.lock = False
360 frac = self._setup_fractionator(
361 simulation_results,
362 purity_required,
(...)
365 allow_empty_fractions=allow_empty_fractions
366 )
--> 368 opt, x0 = self._setup_optimization_problem(
369 frac,
370 purity_required,
371 allow_empty_fractions,
372 ranking,
373 obj_fun,
374 n_objectives,
375 bad_metrics,
376 minimize,
377 )
379 # Lock to enable caching
380 simulation_results.process.lock = True
File ~/checkouts/readthedocs.org/user_builds/cadet-process/conda/latest/lib/python3.11/site-packages/CADETProcess/fractionation/fractionationOptimizer.py:219, in FractionationOptimizer._setup_optimization_problem(self, frac, purity_required, allow_empty_fractions, ranking, obj_fun, minimize, bad_metrics, n_objectives)
216 minimize = False
217 bad_metrics = 0
--> 219 opt.add_objective(
220 obj_fun,
221 requires=frac_evaluator,
222 n_objectives=n_objectives,
223 minimize=minimize,
224 bad_metrics=bad_metrics,
225 )
227 purity = Purity()
228 purity.n_metrics = frac.component_system.n_comp
File ~/checkouts/readthedocs.org/user_builds/cadet-process/conda/latest/lib/python3.11/site-packages/CADETProcess/optimization/optimizationProblem.py:1018, in OptimizationProblem.add_objective(self, objective, name, n_objectives, minimize, bad_metrics, evaluation_objects, labels, requires, *args, **kwargs)
1015 except KeyError as e:
1016 raise CADETProcessError(f"Unknown Evaluator: {str(e)}")
-> 1018 objective = Objective(
1019 objective,
1020 name,
1021 n_objectives=n_objectives,
1022 minimize=minimize,
1023 bad_metrics=bad_metrics,
1024 evaluation_objects=evaluation_objects,
1025 evaluators=evaluators,
1026 labels=labels,
1027 args=args,
1028 kwargs=kwargs
1029 )
1030 self._objectives.append(objective)
File ~/checkouts/readthedocs.org/user_builds/cadet-process/conda/latest/lib/python3.11/site-packages/CADETProcess/optimization/optimizationProblem.py:3863, in Objective.__init__(self, n_objectives, minimize, *args, **kwargs)
3860 def __init__(self, *args, n_objectives=1, minimize=True, **kwargs):
3861 self.minimize = minimize
-> 3863 super().__init__(*args, n_metrics=n_objectives, **kwargs)
File ~/checkouts/readthedocs.org/user_builds/cadet-process/conda/latest/lib/python3.11/site-packages/CADETProcess/optimization/optimizationProblem.py:3762, in Metric.__init__(self, func, name, n_metrics, bad_metrics, evaluation_objects, evaluators, labels, args, kwargs)
3759 self.func = func
3760 self.name = name
-> 3762 self.n_metrics = n_metrics
3764 if np.isscalar(bad_metrics):
3765 bad_metrics = np.tile(bad_metrics, n_metrics)
File ~/checkouts/readthedocs.org/user_builds/cadet-process/conda/latest/lib/python3.11/site-packages/CADETProcess/dataStructure/parameter.py:171, in ParameterBase.__set__(self, instance, value)
169 if value is not None:
170 value = self._prepare(instance, value, recursive=True)
--> 171 self._check(instance, value, recursive=True)
173 try:
174 if self.name in instance._parameters:
File ~/checkouts/readthedocs.org/user_builds/cadet-process/conda/latest/lib/python3.11/site-packages/CADETProcess/dataStructure/parameter.py:483, in Typed._check(self, instance, value, recursive)
480 raise TypeError(f"Expected type {self.ty}, got {type(value)}")
482 if recursive:
--> 483 super()._check(instance, value, recursive)
File ~/checkouts/readthedocs.org/user_builds/cadet-process/conda/latest/lib/python3.11/site-packages/CADETProcess/dataStructure/parameter.py:770, in Ranged._check(self, instance, value, recursive)
756 def _check(self, instance, value, recursive=False):
757 """
758 Validate the value against the range.
759
(...)
768
769 """
--> 770 self.check_range(value)
772 if recursive:
773 super()._check(instance, value, recursive)
File ~/checkouts/readthedocs.org/user_builds/cadet-process/conda/latest/lib/python3.11/site-packages/CADETProcess/dataStructure/parameter.py:752, in Ranged.check_range(self, value)
735 """
736 Validate if the value is within the defined range.
737
(...)
749 If the value is outside the specified bounds.
750 """
751 if self.lb_op(value, self.lb):
--> 752 raise ValueError(f"Value {value} is below the lower bound of {self.lb}")
753 elif self.ub_op(value, self.ub):
754 raise ValueError(f"Value {value} is above the upper bound of {self.ub}")
ValueError: Value False is below the lower bound of 1
The resulting fractionation times show that in this case, it is advantageous to discard some slices of the peak in order not to dilute the overall product fraction.
Exclude Components#
In some situations, not all components are relevant for fractionation. For example, salt used for elution usually does not affect the purity of a component. For this purpose, a subset of components can be specified.
To demonstrate the strategy, consider the LWE example.
Here, the Salt component should not be used for fractionation.
fractionator = fractionation_optimizer.optimize_fractionation(
simulation_results,
components=['A', 'B', 'C'],
purity_required=[0.95, 0.95, 0.95]
)
print(fractionator.performance)
_ = fractionator.plot_fraction_signal()
Sum species#
Note that by default the sum-signal of all component Species is used for fractionation.
To disable this feature, set use_total_concentration_components=False.
For more information on Species, refer to Component System.
